Hopfield¶
-
class
model.hopfield.Hopfield(outputs=100, num_epochs=100, batch_size=100, delta=0.4, optimizer=SGD(lr_max=inf, lr_min=0.0, lr=0.02, decay=0.0), weights_init=Normal(mu=0.0, std=1.0), p=2.0, k=2, precision=1e-30, epochs_for_convergency=None, convergency_atol=0.01, random_state=None, verbose=True)[source]¶ Bases:
plasticity.model._base.BasePlasticityHopfield and Krotov implementation of the BCM algorithm 1.
- Parameters
outputs (int (default=100)) – Number of hidden units
num_epochs (int (default=100)) – Maximum number of epochs for model convergency
batch_size (int (default=10)) – Size of the minibatch
optimizer (Optimizer (default=SGD)) – Optimizer object (derived by the base class Optimizer)
delta (float (default=0.4)) – Strength of the anti-hebbian learning
weights_init (BaseWeights object (default="Normal")) – Weights initialization strategy.
p (float (default=2.)) – Lebesgue norm of the weights
k (int (default=2)) – Ranking parameter, must be integer that is bigger or equal than 2
precision (float (default=1e-30)) – Parameter that controls numerical precision of the weight updates
epochs_for_convergency (int (default=None)) – Number of stable epochs requested for the convergency. If None the training proceeds up to the maximum number of epochs (num_epochs).
convergency_atol (float (default=0.01)) – Absolute tolerance requested for the convergency
random_state (int (default=None)) – Random seed for weights generation
verbose (bool (default=True)) – Turn on/off the verbosity
Examples
>>> from sklearn.datasets import fetch_openml >>> import pylab as plt >>> from plasticity.model import Hopfield >>> >>> X, y = fetch_openml(name='mnist_784', version=1, data_id=None, return_X_y=True) >>> X *= 1. / 255 >>> model = Hopfield(outputs=100, num_epochs=10) >>> model.fit(X) Hopfield(batch_size=100, outputs=100, num_epochs=10, random_state=42, precision=1e-30) >>> >>> # view the memorized weights >>> w = model.weights[0].reshape(28, 28) >>> nc = np.max(np.abs(w)) >>> >>> fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(8, 8)) >>> im = ax.imshow(w, cmap='bwr', vmin=-nc, vmax=nc) >>> fig.colorbar(im, ticks=[np.min(w), 0, np.max(w)]) >>> ax.axis("off") >>> plt.show()
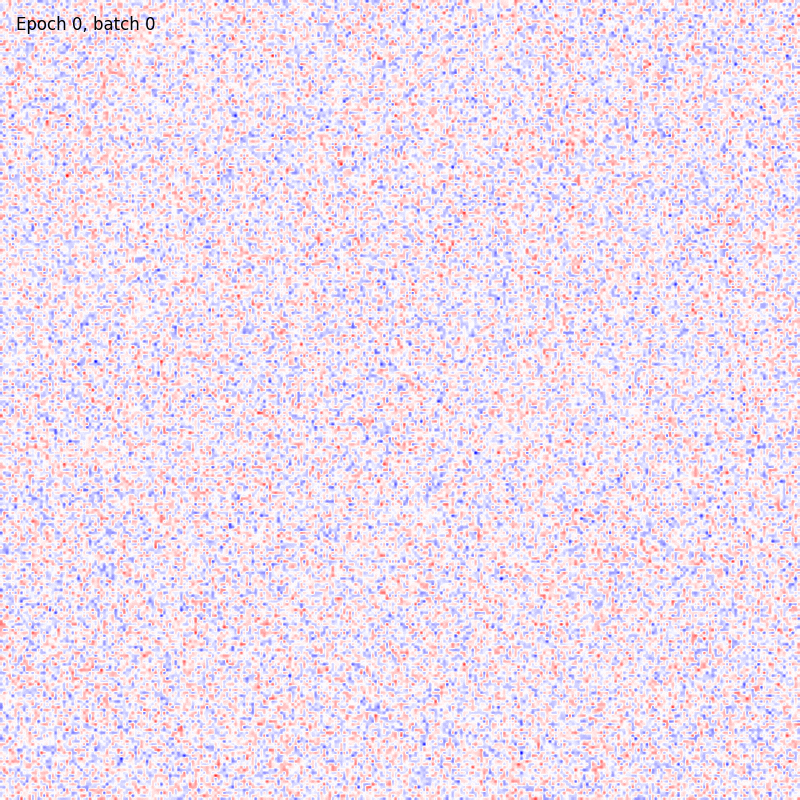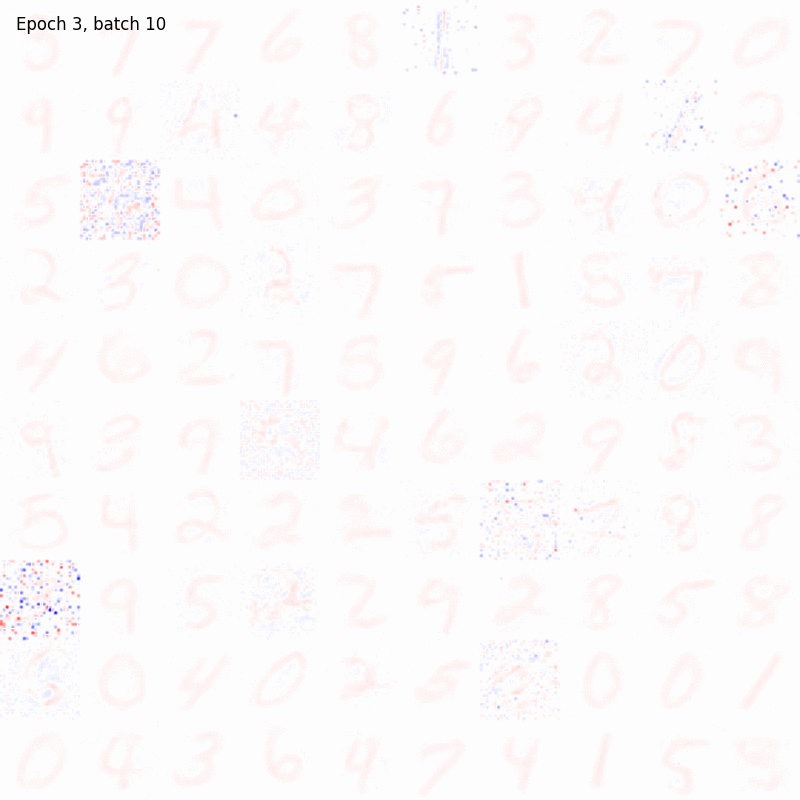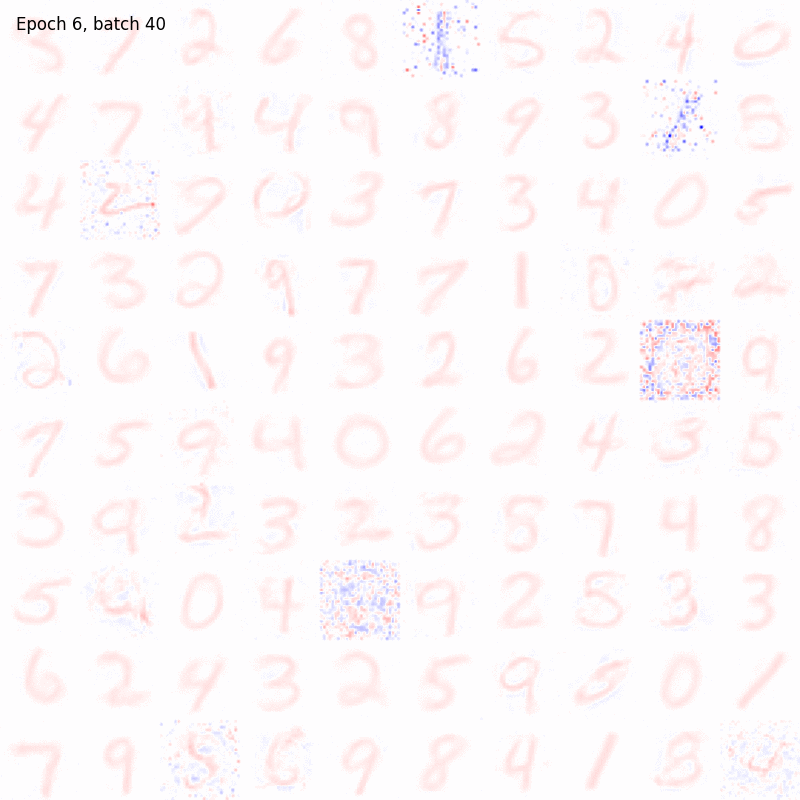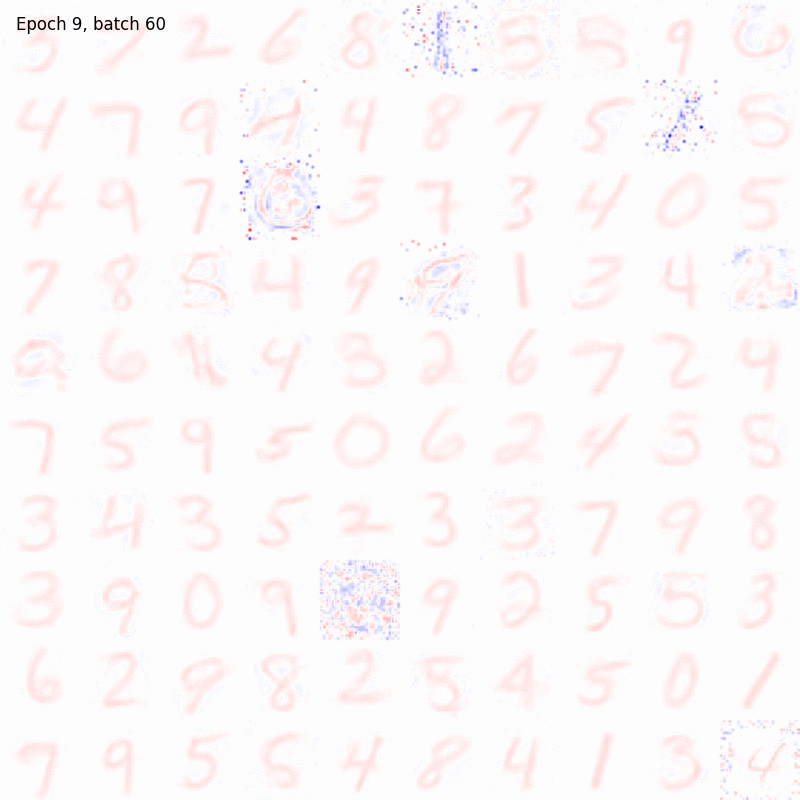
References
- 1
Dmitry Krotov, and John J. Hopfield. Unsupervised learning by competing hidden units, PNAS, 2019, www.pnas.org/cgi/doi/10.1073/pnas.1820458116
-
fit(X, y=None)¶ Fit the Plasticity model weights.
- Parameters
X (array-like of shape (n_samples, n_features)) – The training input samples
y (array-like, default=None) – The array of labels
- Returns
self – Return self
- Return type
object
Notes
Note
The model tries to memorize the given input producing a valid encoding.
Warning
If the array of labels is provided, it will be considered as a set of new inputs for the neurons. The labels can be 1D array or multi-dimensional array: the given shape is internally reshaped according to the required dimensions.
-
fit_transform(X, y=None)¶ Fit the model model meta-transformer and apply the data encoding transformation.
- Parameters
X (array-like of shape (n_samples, n_features)) – The training input samples
y (array-like, shape (n_samples,)) – The target values
- Returns
Xnew – The data encoded according to the model weights.
- Return type
array-like of shape (n_samples, encoded_features)
Notes
Warning
If the array of labels is provided, it will be considered as a set of new inputs for the neurons. The labels can be 1D array or multi-dimensional array: the given shape is internally reshaped according to the required dimensions.
-
get_params(deep=True)¶ Get parameters for this estimator.
- Parameters
deep (bool, default=True) – If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
params – Parameter names mapped to their values.
- Return type
mapping of string to any
-
load_weights(filename)¶ Load the weight matrix from a binary file.
- Parameters
filename (str) – Filename or path
- Returns
self – Return self
- Return type
object
-
predict(X, y=None)¶ Reduce X applying the Plasticity encoding.
- Parameters
X (array of shape (n_samples, n_features)) – The input samples
y (array-like, default=None) – The array of labels
- Returns
Xnew – The encoded features
- Return type
array of shape (n_values, n_samples)
Notes
Warning
If the array of labels is provided, it will be considered as a set of new inputs for the neurons. The labels can be 1D array or multi-dimensional array: the given shape is internally reshaped according to the required dimensions.
-
save_weights(filename)¶ Save the current weights to a binary file.
- Parameters
filename (str) – Filename or path
- Returns
- Return type
True if everything is ok
-
set_params(**params)¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
**params (dict) – Estimator parameters.
- Returns
self – Estimator instance.
- Return type
object
-
transform(X)¶ Apply the data reduction according to the features in the best signature found.
- Parameters
X (array-like of shape (n_samples, n_features)) – The input samples
- Returns
Xnew – The data encoded according to the model weights.
- Return type
array-like of shape (n_samples, encoded_features)