BCM¶
-
class
model.bcm.BCM(outputs=100, num_epochs=100, batch_size=100, activation='Logistic', optimizer=SGD(lr_max=inf, lr_min=0.0, lr=0.02, decay=0.0), weights_init=Normal(mu=0.0, std=1.0), interaction_strength=0.0, precision=1e-30, epochs_for_convergency=None, convergency_atol=0.01, random_state=None, verbose=True)[source]¶ Bases:
plasticity.model._base.BasePlasticityBienenstock, Cooper and Munro algorithm (BCM) 1.
The idea of BCM theory is that for a random sequence of input patterns a synapse is learning to differentiate between those stimuli that excite the postsynaptic neuron strongly and those stimuli that excite that neuron weakly. Learned BCM feature detectors cannot, however, be simply used as the lowest layer of a feedforward network so that the entire network is competitive to a network of the same size trained with backpropagation algorithm end-to-end.
- Parameters
outputs (int (default=100)) – Number of hidden units
num_epochs (int (default=100)) – Maximum number of epochs for model convergency
batch_size (int (default=10)) – Size of the minibatch
activation (string or Activations object (default='Logistic')) – Activation function to apply
optimizer (Optimizer (default=SGD)) – Optimizer object (derived by the base class Optimizer)
weights_init (BaseWeights object (default="Normal")) – Weights initialization strategy.
interaction_strength (float (default=0.)) – Set the lateral interaction strenght between weights
precision (float (default=1e-30)) – Parameter that controls numerical precision of the weight updates
epochs_for_convergency (int (default=None)) – Number of stable epochs requested for the convergency. If None the training proceeds up to the maximum number of epochs (num_epochs).
convergency_atol (float (default=0.01)) – Absolute tolerance requested for the convergency
random_state (int (default=None)) – Random seed for weights generation
verbose (bool (default=True)) – Turn on/off the verbosity
Examples
>>> from sklearn.datasets import fetch_openml >>> import pylab as plt >>> from plasticity.model import BCM >>> >>> X, y = fetch_openml(name='mnist_784', version=1, data_id=None, return_X_y=True) >>> X *= 1. / 255 >>> model = BCM(outputs=100, num_epochs=10) >>> model.fit(X) BCM(batch_size=100, outputs=100, num_epochs=10, random_state=42, epsilon=0.02, precision=1e-30) >>> >>> # view the memorized weights >>> w = model.weights[0].reshape(28, 28) >>> nc = np.max(np.abs(w)) >>> >>> fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(8, 8)) >>> im = ax.imshow(w, cmap='bwr', vmin=-nc, vmax=nc) >>> fig.colorbar(im, ticks=[np.min(w), 0, np.max(w)]) >>> ax.axis("off") >>> plt.show()
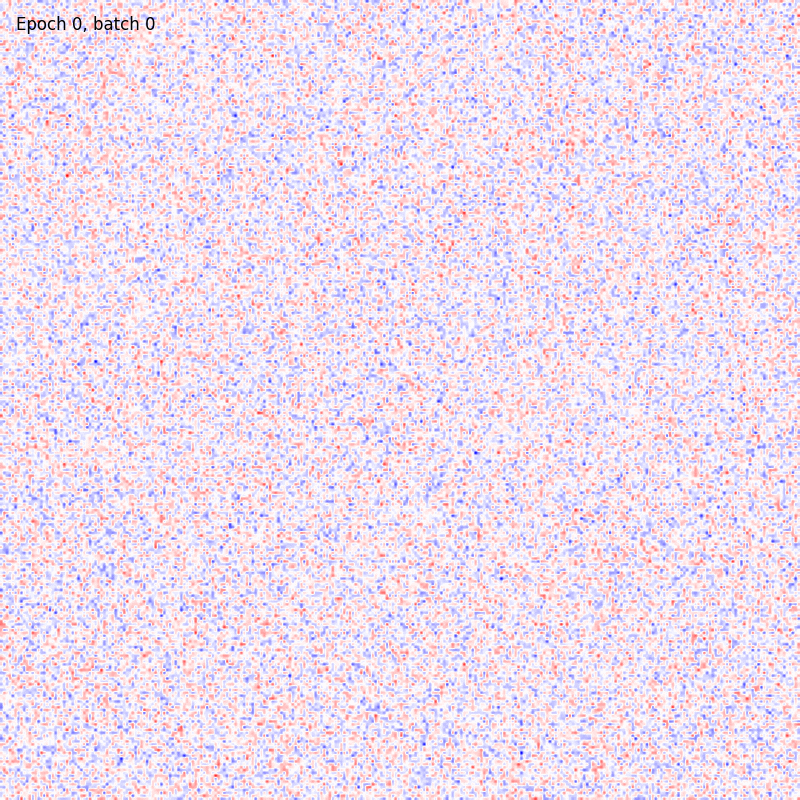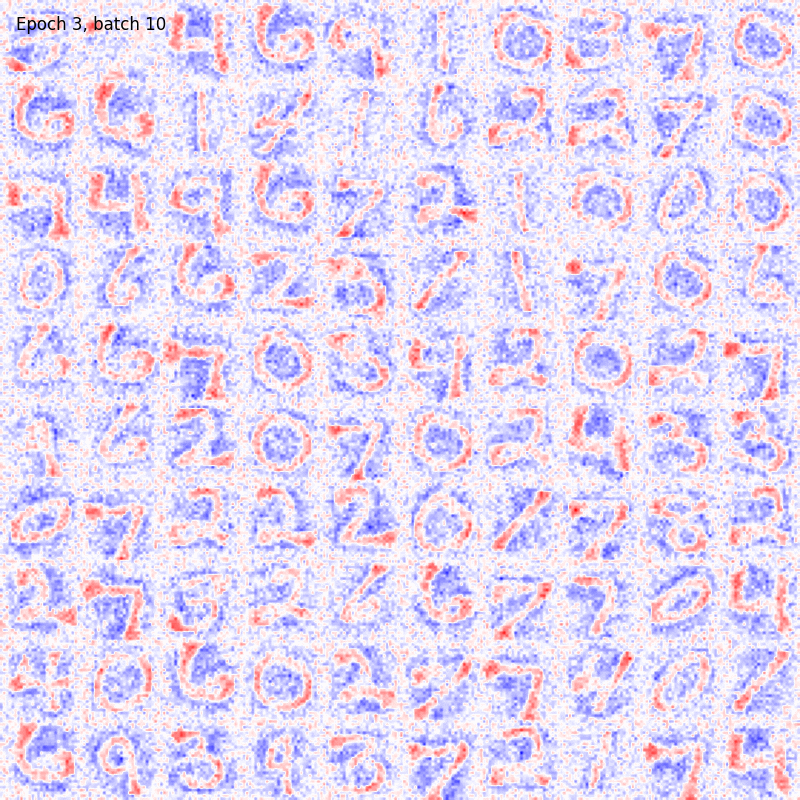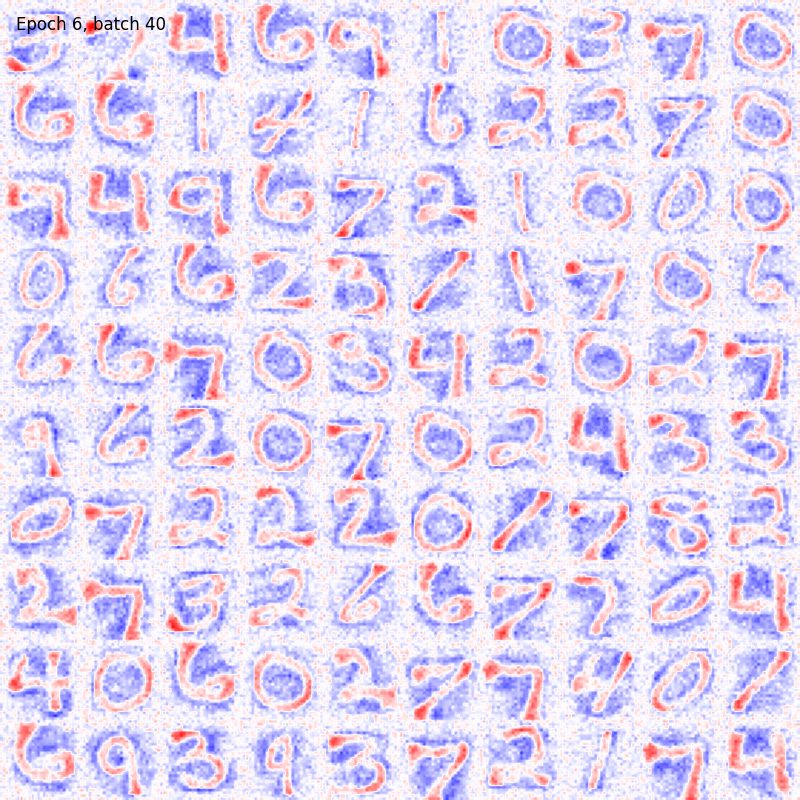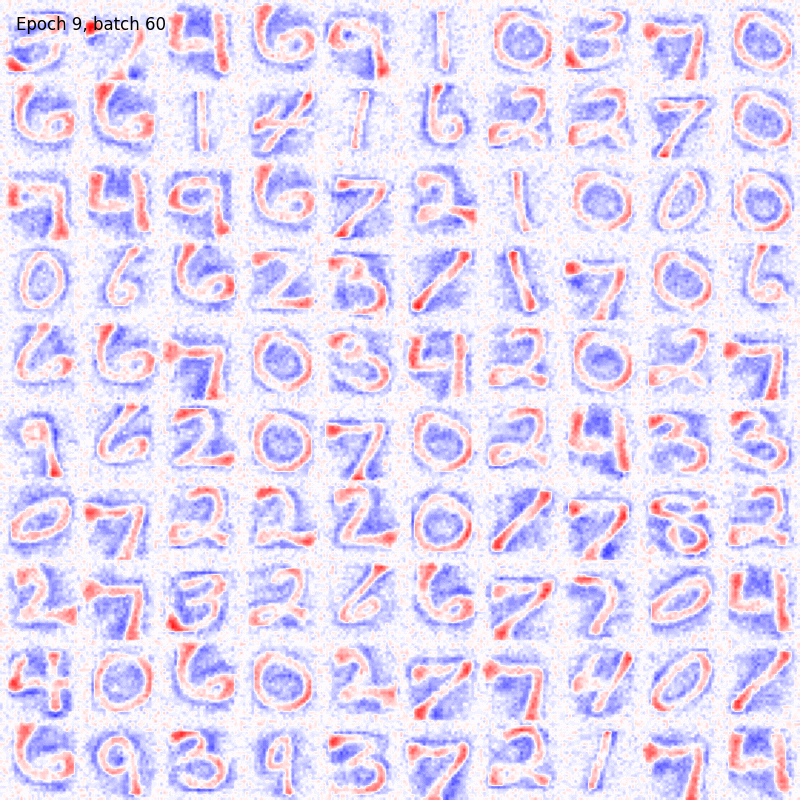
References
- 1
Castellani G., Intrator N., Shouval H.Z., Cooper L.N. Solutions of the BCM learning rule in a network of lateral interacting nonlinear neurons, Network Computation in Neural Systems, 10.1088/0954-898X/10/2/001
-
fit(X, y=None)¶ Fit the Plasticity model weights.
- Parameters
X (array-like of shape (n_samples, n_features)) – The training input samples
y (array-like, default=None) – The array of labels
- Returns
self – Return self
- Return type
object
Notes
Note
The model tries to memorize the given input producing a valid encoding.
Warning
If the array of labels is provided, it will be considered as a set of new inputs for the neurons. The labels can be 1D array or multi-dimensional array: the given shape is internally reshaped according to the required dimensions.
-
fit_transform(X, y=None)¶ Fit the model model meta-transformer and apply the data encoding transformation.
- Parameters
X (array-like of shape (n_samples, n_features)) – The training input samples
y (array-like, shape (n_samples,)) – The target values
- Returns
Xnew – The data encoded according to the model weights.
- Return type
array-like of shape (n_samples, encoded_features)
Notes
Warning
If the array of labels is provided, it will be considered as a set of new inputs for the neurons. The labels can be 1D array or multi-dimensional array: the given shape is internally reshaped according to the required dimensions.
-
get_params(deep=True)¶ Get parameters for this estimator.
- Parameters
deep (bool, default=True) – If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
params – Parameter names mapped to their values.
- Return type
mapping of string to any
-
load_weights(filename)¶ Load the weight matrix from a binary file.
- Parameters
filename (str) – Filename or path
- Returns
self – Return self
- Return type
object
-
predict(X, y=None)¶ Reduce X applying the Plasticity encoding.
- Parameters
X (array of shape (n_samples, n_features)) – The input samples
y (array-like, default=None) – The array of labels
- Returns
Xnew – The encoded features
- Return type
array of shape (n_values, n_samples)
Notes
Warning
If the array of labels is provided, it will be considered as a set of new inputs for the neurons. The labels can be 1D array or multi-dimensional array: the given shape is internally reshaped according to the required dimensions.
-
save_weights(filename)¶ Save the current weights to a binary file.
- Parameters
filename (str) – Filename or path
- Returns
- Return type
True if everything is ok
-
set_params(**params)¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
**params (dict) – Estimator parameters.
- Returns
self – Estimator instance.
- Return type
object
-
transform(X)¶ Apply the data reduction according to the features in the best signature found.
- Parameters
X (array-like of shape (n_samples, n_features)) – The input samples
- Returns
Xnew – The data encoded according to the model weights.
- Return type
array-like of shape (n_samples, encoded_features)